2.2 – Why do we use R Software?
- Why R? The case for using R in biostatistics education.
- How to get started with R .
- Why R Commander, why not RStudio? .
- Wait! Why don’t we use Microsoft Excel? .
- Statistics comparisons between R and MS Excel .
- Graphics comparison between R and MS Excel .
- So, you’re telling me I don’t need a spreadsheet application? .
- Still not convinced? .
- Why install R on your computer?.
- Help with R?.
- Questions .
- Chapter 2 contents.
Why R? The case for using R in biostatistics education
.
Why do we use R Software? Or put another way: Dr D, Why are you making me use R?
Truth? You can probably use just about any acceptable statistical application to get the work done and achieve the learning objectives we have for beginning biostatistics. However, we will use the R statistical language as our primary statistical software in this course. Part of the justification is that all statistical software applications come with a learning curve, so you’d start at zero regardless of which application I used for the course. In selecting software for statistics I have several criteria. The software should be:
- if not exactly easy, the software should have a reasonable learning curve
- widely accessible and compatible with
allmost personal computers - well-respected and widely used by professionals
- free software
- open source
- well-supported for the purposes of data analysis and data processing
- really good for making graphics, from the basics to advanced
- capable to handle diverse kinds of statistical tests
R meets all of these criteria. R history began back in 1993 and has always been available as free software under the terms of the Free Software Foundation’s GNU General Public License in source code form. R compiles and runs on a wide variety of UNIX platforms and similar systems, including GNU/LINUX, FreeBSD, and various Linux distros like the popular Ubuntu®, in addition to their more famous Microsoft Windows® and Apple macOS® distributions. To facilitate access to the software, numerous mirror sites are available from sites around the world, with cloud.r-project.org supported by RStudio perhaps the most widely used. From January 2023 to December 2023, more than 8 million downloads of base R were made from the RStudio CRAN mirror site (CRAN stands for Comprehensive R Archive Network; a mirror refers to a website or server that holds a copy of files from another website/server to make the files available from more than one place).
changelog file, which allow one to track numbers of downloads for any package from their mirror site — https://cloud.r-project.org/. Here’s the code and recent counts for downloads of R itself (about 400K over a four week period).
install.packages("cranlogs")
library(cranlogs)
# How many downloads of base R first four weeks of Fall semester?
out <- cran_downloads("R", from = "2023-09-04", to = "2023-09-29")
sum(out$count)
R output
[1] 887417
R is straight-forward to use once you learn how to work with the language, but has a steep learning curve; after all, it’s a programming language. The GUI R Commander helps in this process, and eventually, your use of code will become second nature. After the initial growing pains are behind you, RStudio likely will be a better solution over R Commander. However, while we need statistical software to do statistics, students in my BI311 course must keep in mind that learning objectives for most biostatistics course are about the concepts and interpretation of statistics, not just use of the software. In other words, learning how to use R is not the focus of BI311 nor will you likely achieve R programming competency by the end of the semester. I certainly encourage students to strive for competency and I give frequent bonus opportunities to demonstrate coding skills during the semester.
Thus you might ask if the purpose of the course isn’t to learn R, why work with R instead of a more familiar app or software, e.g., Microsoft Excel® (hereafter simply referred to as Excel), or Google Sheets, or even my favorite open-source office alternative, LibreOffice Calc? Or, perhaps even just one of the many online calculators, if the course learning objective is to “just” learn about statistics?
First, I believe that real data derived from real biology or biomedical problems are essential elements to a first course in biostatistics. That’s not a particularly unique perspective although I don’t have survey results of other statistics instructors to back up the claim. Real problems involve observations on multiple subjects, many variables — large data sets; this alone precludes use of hand calculations and calculators. As a corollary, we will not spend a great deal of time learning the in’s and out’s of the algorithms that form particular statistical tests. Now, do understand that there is a tremendous benefit to understanding statistics by working through the equations, by looking at the algorithms, and there’s no escaping the need for understanding that probability provides the foundation of statistics inference (Chapter 8). Thus, for most of us, the statistical software available to us provides an appropriate framework for applying correct statistical tests to our projects. Therefore, the decision is about which statistical package we should use.
Second, R is perhaps the choice in academia for statistical software. A PUBMED search found more than 1500 citations of R. Visit Robert A. Muenchen’s web page (The popularity of data analysis software, r4stats.com) to see updated statistics on statistical software use. Those of you continuing on to graduate school or to professional schools will find that many of your statistically literate colleagues use R and not one of the commercial programs. While there are many excellent commercial packages (Table 1), and in some cases you can make spreadsheet programs do statistics (typically add-ins are required), all statistical software come with steep learning curves. Thus, part of my selling point to you is that learning to use R is at the cutting-edge in your field and, given that all of the software you could use can have have their challenges, it is best to work with something that will be around and is in wide use, without the burden of a financial investment.
Table 1. A selective list of statistical software.
| Software | Student license? | Limited or full function version | macOS | Windows 11 | Fee* | Academic license type |
|---|---|---|---|---|---|---|
| GraphPad Prism | Subscription, $142 per year | Full | Yes | Yes | $202 | annual subscription |
| JMP | Yes, but with purchase of selected textbook | Limited | Yes | Yes | $100 | monthly subscription |
| Minitab | Subscription, $54.99 per year | Full | Yes | Yes | $1610 | annual subscription |
| IBM SPSS | Rental, $76 per year | Full | Yes | Yes | $260 | annual rental |
| SigmaSTAT | No | NA | No | Yes | $299 | perpetual |
| MySTAT | Yes, free | Limited | No | Yes | NA | |
| SYSTAT | No | NA | No | Yes | $739 | perpetual |
| Stata | Subscription, $94 per year | Full | Yes | Yes | $325 | annual subscription |
see Wikipedia for list of additional software
Third, what about online sites like plot.ly where, for free, you can plot and, in some cases, calculate statistics? What about the web application at Brightstat, which claims to provide an SPSS-like experience online (Stricker 2008)? While it is true that there are many wonderful websites that can perform many of the statistical tests we will use this semester, these sites are not suitable for more than occasional use.
How to get started with R
.
First, a few words about my approach to the software in my biostatistics course. I note to students during the semester that our course is not a programming course, but rather, programming skills are acquired along the way. Our focus is how to do statistical analysis, how to design experiments, etc. Coding skills without understanding of the why we’re doing it is not unlike learning to cook by following a great recipe as opposed to learning about the art and science of flavor (NPR Science Friday, March 15, 2024) — the homework is solved but the why we can support our conclusions may be obscure.
We set aside the first couple of class meetings simply to setup student’s Windows PC or Apple macOS computers to do data science; for students with access to Chromebooks or iPad tablets, we need to set them up to run R in the Cloud (eg., Google Colaboratory) — in other words, there’s a lot of pre-work that can present barriers to actual doing statistics in R (or other software). Once we have the environment set up for the students, I utilize a “tell-show-do” approach to learning how to code. The “tell” part includes sharing script; the “do” part is mostly done by students outside of class as part of completing homework. First work with the language can be frustrating, so I also encourage a “five minute” rule — if it ain’t working, stop. So, by all means when Run through a troubleshooting checklist, but please don’t continue to struggle — the gap is between the instructions and where the student is with the material — it’s a me as instructor problem, not a student must work harder problem!
The R statistical language, accompanied by additional packages to extend its capabilities beyond basic math and statistical functions, provided a complete statistical environment. R is best viewed as a programming language for statistics (data analysis), and data processing. Power users of R learn how to write scripts that do t-tests, ANOVA, regression, etc. The scripts are just lines of code that R understands and it provides the user tremendous control over analysis and inference of data sets. Because of this flexibility and power, however, R can be intimidating at first. So, we’ll start slowly with scripts, introducing just what we need to get started and build from there. We’ll be addressing R issues in more depth over the next several weeks, but for the first week, our goal(s) should be to make sure each of you knows how to start/exit R, how to create and utilize a working directory, and how to use R as a calculator. You obtain your copy of R from the R Project for Statistical Computing, available at https://www.r-project.org. Instructions to install R are provided in Install R. A ten-part tutorial to get started using R is provided in Mike’s Workbook for Biostatistics.
Note 2. A working directory or working folder is something you create on your computer to contain the files and sub-directories of a project. It sets the default location for files you may need to have R read. For example, all of your work for a course (data files, script files, Markdown files), may be stored in a folder called BI311 on your Desktop. For example, on a macOS, the path to the working folder would be
/Users/username/Desktop/BI311
Why R Commander, why not RStudio?
.
We utilize an R package that provides a menu-driven context to much of the typical statistics one needs to do biostatistics. The package is called R Commander (Rcmdr), which provides a graphic user interface or GUI. Rcmdr therefore significantly eases the learning curve for doing statistics with R. We use a package called R Commander, which provides drop down menus for most of the typical kinds of analyses. Rcmdr is in use in many courses across the world (more than 20K downloads in September 2023), and among the other GUI available for R, Rcmdr is among the best supported GUI available for R. R Commander function is extended by plug-ins; as of August 2023, there were 36 plugins that extend Rcmdr’s capabilities. Instructions to install R Commander are provided in Install R Commander.
Note 3. Other options to improve use of R include use of RStudio®, which is an integrated development environment or IDE. RStudio is really nice to use, and, happily, you can run R Commander within RStudio — but with windows popping up outside of the RStudio windowing framework if the default MDI environment to organize is set. (The solution? R Commander runs best when SDI option is selected during RStudio install.) I am also increasingly using shiny apps within the course to help with concept presentation; in the future, I plan to provide a complete shiny app which would allow BI311 students to work interactively with the statistics presented in this text, something like the radiant-rstats project. However, for use in our course, R Commander provides a familiar look as students develop knowledge in the course: simply point and click to access the statistical functions.
Wait! Why don’t we use Excel? My instructor in {insert course here} used Excel…
.
A very reasonable question for you to ask — why don’t we use Microsoft Excel or Google Sheets for statistics? Moreover, it is highly likely that you have gained at least some introduction to descriptive statistics and graphing with spreadsheets in former courses — shouldn’t we learn statistics within a software framework you are already familiar?
After all, “Can’t Microsoft Excel do statistics?” Mostly the answer is, no, not really (Fig 1).
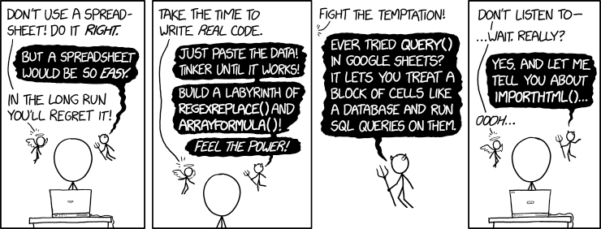
Figure 1. “Spreadsheets,” xkcd.com no. 2180
MS Excel, Google Sheets, Apple Numbers, and for that matter, Calc, the spreadsheet application in my favorite office app LibreOffice (LibreOffice is a free, open-source alternative to Microsoft Office), can be used to calculate many descriptive statistics. With some effort, these applications can be extended by use of either Analysis ToolPak or Solver Add-ins to do more complicated statistics like regression and analysis of variance, and curve fitting.
Note 4. Perhaps you’re thinking: I’m a data science student — it’s not whether we chose between use of spreadsheet apps or R, we should be using Python, shouldn’t we? DrD — this is an open question, and if you judge the job market as the arbiter, I’d say learn Python over R. However, from the point of view of learning statistics, my vote would be R — far more packages developed to accomplish our tasks, and, at least for this course, we are not teaching coding: our focus is on using statistics to explore (describe) and ask questions about data collected by observation or experimentation.
While we’re on the subject in 2025, it looks like the job market is in Machine Learning, which means R or Python knowledge will be a given, but advantage will go to those who know Alteryx or it’s open source alternative KNIME for data science workflow design — from data collection to model development.
However, use of MS Excel for statistical analysis involves learning a number of commands, syntax, and developing workflows that are neither intuitive nor standard. Some publishers have provided add-ins that are reportedly designed to simplify this process (eg, XLStat, UNISTAT, Real Statistics using Excel). None of these options are free and none are in use in any major way by scientists (see Robert Muenchen’s The popularity of data analysis software). The free add-ins of Analysis ToolPak and Solver may work for you if you own a Windows PC, but only Solver is included for the Mac versions of Excel. MacOS users may download and install StatPlus:MacLE, which is a limited, but free alternative to the Analysis ToolPak add-in; for a complete package a Pro version is available (licenses started at $89, web site: www.analystsoft.com/en/products/statplusmacle/).
An additional caution: you should be aware that there have been reports over the years that algorithms selected by Microsoft for Excel have not always been to industry standards (e.g., McCullogh and Wilson 2005). In short, the fit of Excel and other spreadsheet apps for use in statistics is not a simple one. To do the kinds of statistics we will use routinely in class, Excel would need to be modified with add-ins, and the add-ins would be the result of programming by someone. And you would still need to learn how to write the code.
What about graphics? You may like Microsoft Excel’s ability to do graphics. Indeed, Excel, Google Sheets, and LibreOffice Calc can be used to generate many typical kinds of statistical plots. But again, in comparison to R, spreadsheet app graphics are limited and require a deal of effort to generate acceptable plots. I think you’ll be surprised at how straight-forward R is. Here’s an example, first rendered in Microsoft Excel, then in base R. And importantly, the kinds of plots Excel does well at are not necessarily the plots suitable for research publication. For example, Excel allows you to make bar charts easily, but cannot do box plots. Box plots are preferred over bar (column) charts for ratio scale data.
Note 5. base R refers to the core R programming language along with many functions and graphics routines. We extend capabilities of base R by adding packages, like R Commander.
Statistics comparisons between R and MS Excel
.
About that learning curve. Let’s compare R and MS Excel for basic functions common in data analysis. Similar conclusions hold for comparisons to Google Sheets and LibreOffice Calcs.
Note 6. If you search for variants of this question, you’ll find other’s making the argument that the learning curve for many data science tasks is, perhaps surprisingly, shorter for R than it is for Excel. For example, see Amieroh Abrahams 2023 article at https://www.jumpingrivers.com/blog/comparing-r-excel-data-wrangling/ .
Table 2 lists the observations we can use to conduct comparisons of the applications.
Table 2. A simple data set, one variable, A, with 24 observations
| varA |
|---|
| 12 |
| 14 |
| 20 |
| 25 |
| 28 |
| 29 |
| 32 |
| 34 |
| 35 |
| 39 |
| 47 |
| 47 |
| 50 |
| 53 |
| 54 |
| 71 |
| 79 |
| 87 |
| 89 |
| 96 |
| 105 |
| 122 |
| 130 |
| 132 |
One of the first steps in data analysis is to produce what are called descriptive statistics. Common descriptive statistics are the mean and the sample standard deviation. Let’s compare Excel and R for retrieving these two statistics.
With Excel, to calculate the arithmetic mean of 24 numbers, enter the values into a single column of 24 rows, then enter “=average(A2:A25)“, without the quotes, into a new cell of the spreadsheet. “A2:A25” refers to where data would be contained in column A rows 2 through 25. Typically the first row in a worksheet would contain the name of the variable, e.g., “A.” Depending on the significant figures set, the estimate returned by Excel for the mean of A is 59.58333333.
Similarly, to obtain the standard deviation, type =stdev(A2:A25), into a new cell of the spreadsheet. Again, depending on the significant figures set, Excel returns a value of 37.05215674 for the standard deviation of A.
In contrast, to obtain the mean and standard deviation for a variable in an R data set, all you would type at the R prompt (>), or in the script window
Note 7. Always run your code as a script. Entering code at the R prompt means you are working at the command-line interface, and you work one line at a time. This is not an efficient way to interact with R. Instead, I recommend you always create and work from a script document. For beginners, that’s why I recommend R Commander, which includes a script window. Simply type your code in the script window, highlight the code you wish to run, and run by clicking submit button (or Ctrl+R Win11 or Cmd+Enter macOS). When you are ready to move on from R Commander, RStudio is the IDE of choice.
and then submit the code is:
A <- c(12, 14, 20, 25, 28, 29, 32, 34, 35, 39, 47, 47, 50, 53, 54, 71, 79, 87, 89, 96, 105, 122, 130, 132)where the “c” is a function to combine arguments into a vector and saved to the object A, followed at the new line by
mean(A)Hit enter after entering the command) and R returns
[1] 59.58333For the standard deviation, write the R base function sd()
sd(A)Hit enter after entering the command and R returns
[1] 37.05216It’s not much of a difference, but note that to get the mean (arithmetic average) I typed seven characters in R, but 16 characters in Excel; similarly, for the standard deviation I typed in 5 characters in R, but 13 characters in Excel. That’s a savings of 56% and 62%, respectively. Excel tries to help by using AutoComplete to anticipate what you want to enter, but AutoComplete doesn’t always work properly (e.g., see gene name errors generated by use of default Microsoft Excel settings, Ziemann et al 2016).
Note 8. I use spreadsheets all of the time for data entry and data management. Make sure AutoComplete and AutoCorrect options are turned off and these problems are much less.
In conclusion, R is quicker for descriptive statistics.
Graphics comparison between R and MS Excel
.
MS Excel is often cited for its graphics capabilities (Camões 2016). We can make the familiar scatter plots, bar charts, and pie charts in Excel. These plots and more are easily obtained in R. I won’t elaborate here about graphics, we talk at some length about graphics in Chapter 4. But here’s one example in R.
Let’s plot B vs A. We already provided the data for variable A, here’s the data for variable B.
17, 21, 21, 26, 27, 32, 28, 42, 40, 30, 71, 53, 56, 61, 55, 89, 82, 63, 116, 162, 116, 154, 137, 149Don’t recall how to assign a set of numbers to an object, B, in R? See above and look again at how we assigned the numbers to object A.
To get a simple scatter plot (Fig 2), I may write at the R prompt.
plot(A,B)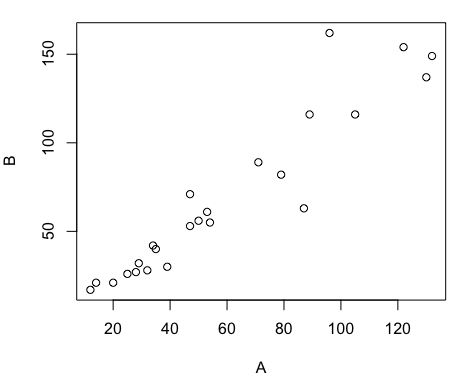
Figure 2. Basic scatter plot made in R, plot(A,B).
And here’s the comparable default plot (Fig 3) from Microsoft Excel, Office 365
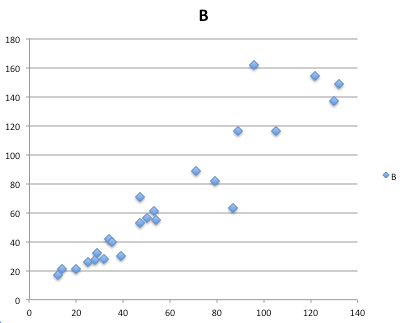
Figure 3. Basic scatterplot made in Microsoft Excel.
Now, both graphs need some work, and to be fair, these are just the defaults. With some effort, you can make an Excel graph look pretty good. But note — the defaults in Excel don’t generate axis labels, while R default plot does. Excel adds a useless title and legend; both need to be removed. Excel also adds grid lines where typically one would not include these in a scientific plot.
Count the steps to generate an acceptable scatter plot (Table 3). I’ve also added R Commander (Rcmdr) steps for comparisons (Rcmdr lets you use drop-down menus like Excel or Google Sheets or LibreOffice Calcs).
Table 3. Steps needed to make a simple scatterplot in R, R Commander, or Microsoft Excel.
| Steps | R | Rcmdr | Excel 365 |
| 1 | write the function | Select Graphs | Highlight columns |
| 2 | Select scatterplot | Select from Menu “Insert” | |
| 3 | Select variables | Select scatterplot | |
| 4 | Uncheck options | Select type of scatterplot | |
| 5 | Delete legend | ||
| 6 | Remove grids | ||
| 7 | Insert X-axis label | ||
| 8 | Insert Y-axis label |
Conclusion? R is quicker for routine statistical plots like a scatter plot. And I didn’t even count the steps needed to change MS Excel’s dreadful diamond icon points.
That’s one step in R, four steps in Rcmdr, but eight steps for Microsoft Excel. LibreOffice Calc is a little better at four steps, but like MS Excel, you’d need to change several components to the graph (Fig 4).
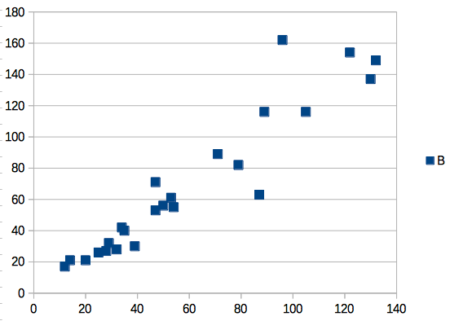
Figure 4. Basic scatterplot made in LibreOffice Calc.
Note 9. In R vernacular, these are referred to as pch, or point characters: pch = 23 returns a blue diamond character; for a blue square like Figure 4, add to the plot() command as
plot(A,B, pch = 22)
So, you’re telling me I don’t need a spreadsheet application?
.
No, not at all. We use spreadsheets, and more generally, databases, to store data. Spreadsheets apps are designed to make data entry and data management approachable and efficient. They remain an important tool for researchers (Browman and Woo 2017).
R is not that great of a spreadsheet; packages are available to seamlessly tie your spreadsheet and database data to R via ODBC. We will routinely enter and manipulate data in MS Excel, then import the data into R for analysis.
Spreadsheet apps like MS Excel and Google Sheets (see also LibreOffice Calc) are great at being a spreadsheet program, R is great at being a statistical software program. You should take advantage of what the tools do best.
Still not convinced?
.
R is in use all over the world, by students and professionals alike and if one is going to spend the time to learn how to use a statistics software program, you should learn a standard program, like R.
And it’s not just me. Read about R in this 2009 New York Times piece, “Data analysts captivated by R’s power.” Look who purchased (April 2015) Revolution Analytics, a major player in the development of the R programming language.
Note 10. The answer was Microsoft. For several years Microsoft supported R development via Microsoft Machine Learning Server & Microsoft R Open. However, as of July 2023, this service is no longer available. See Microsoft R application network retirement.
Why install R on your computer?
.
Convenience. Control. Offline.
At the Biology department of Chaminade University, we have installed and maintain R, Rcmdr, and RStudio along with all required packages on our Macbook Pro® Lab computers for your use during class and during optional, proctored biostatistics work sessions. Since 2018, R is increasingly available “in the cloud” (e.g., RStudio Cloud), which would mean you could run R in your browser and avoid installation on your computer. You can run significant analysis with R in the cloud via the free Google Colaboratory and CoCalc are now available: I encourage you to look into these platforms. Unfortunately, these services are not quite ready for the classroom. For example, RStudio in the Cloud is free to use on a limited basis, but quickly requires a significant subscription cost with increasing use. Google Colab and GoCalcs require use of Jupyter notebooks, which add yet another layer to the learning curve without focusing on learning statistics. Second, although access to their servers is easy, running simultaneous connections via Chaminade’s single public IP address is likely to lead to problems for us. Third, I want you to use R Commander (Rcmdr) to assist in the learning curve — Rcmdr cannot be run in the Cloud (i.e., RStudio in the Cloud, Google Colaboratory, or CoCalc).
Therefore, you are encouraged to install R, Rcmdr, and even RStudio, onto your own computers, in part because of the convenience, but also because R is not generally available to students on campus, i.e., only the Biology department’s computers have the up-to-date R software installed.
To get started, go to your Canvas website and view How to install R on your own computer.
An additional benefit to installing a version of R on your computer, you’ll understand more about the software if you take the time to install and if need be, troubleshoot your installation of the software. Moreover, there’s a considerable amount of help out there for R. For example, a simple Google search(keywords: tutorial “install R”), returns more than 700K hits, and more than 40K January 2023 alone (add “after:2023-01-01” to Google search box). In fact, there’s so much out there that you’ll want to sample from several sites and select the voice that works best for you.
Questions
.
- Conduct the search on Google for tutorials on installing R; find 10 sites and rank them 1 to 10, with 1 being the site you like best and 10 being the one you like least.
- For example, I like https://bookdown.org/ndphillips/YaRrr/, which is an online book for working with R and includes detailed instructions for installing R.
- What are the three reasons I offered to justify use of R over other candidate statistical applications?
- R may be installed on the public computers available to you in the lab. Check to see if this is true, and if so, what version of R is installed?
- What does Rcmdr stand for?
- In your own words, define and contrast GUI applications from IDE applications
- Try some R work yourself
- In R (or Rcmdr), copy and paste the code above for the
Avariable, then create theBvariable. What happens when you type the variable name by itself at the R prompt? - Make a plot of
AandB, but this time plotAagainstB.- What can you conclude about the axis order in the function?
- In R (or Rcmdr), copy and paste the code above for the
Chapter 2 contents
.
1.1 – A quick look at R and R Commander
A first look at R
.
R is a programming language for statistical computing (Venables et al 2009). R is an interpreted language — when you run (execute) an R program — the R interpreter intercepts and runs the code immediately through its command-line interface, one line at time. Python is another popular interpreted language common in data science. Interpreted languages are in contrast to compiled languages, like C++ and Rust, where program code is sent to a compiler to a machine language application.
The following steps user through use of R and R Commander, from installation to writing and running commands. Mike’s Workbook for Biostatistics has a ten-part tutorial, A quick look at R and R Commander, which I recommend.
Note: Getting started? By all means rely on Mike’s Biostatistics Book and blogs or other online tutorials to point you in the right direction. You’ll also find many free and online books that may provide the right voice to get you working with R. However, the best way to learn is to go to the source. The R team has provided extensive documentation, all included as part of your installation of R. In R, run the command RShowDoc("doc name"). replace doc name with the name of the R manual or R user guide. For example, the Venables publication is accessed as RShowDoc("R-intro"). Similarly, the manual for installation is RShowDoc("R-admin") and the manual for R data import/export is RShowDoc("R-data").
Install R
.
Full installation instructions are available at Install R, and for the R Commander package, at Install R Commander. Here, we provide a brief overview of the installation process.
Note: The instructions at Mike’s Biostatistics Book assume use of R on a personal computer running updated Microsoft Windows or Apple macOS operating systems. For Linux instructions, e.g., Ubuntu distro, see How to install R on Ubuntu 22.04. For Chromebook users, if you can install a Linux subsystem, then you can also install and run R although it’s not a trivial installation. For instructions to install R see Levi’s excellent writeup at levente.littvay.hu/chromebook/. (This works best with Intel-based CPUs — see my initial attempts with an inexpensive Chromebook at Install R.)
Another option is to run R in the cloud via service like Google’s Colab or CoCalc hosted by SageMath. Both support Jupyter Notebooks, a “web-based interactive computational environment.” Neither cloud-based service supports use of R Commander (because R Commander interacts with your local hardware). Colab is the route I’d choose if I don’t have access to a local installation of R. See Use R in the cloud for more details.
Download a copy of the R installation file appropriate for your computer from one of the Comprehensive R Archive Network (CRAN) mirror site of the r-project.org. For Hawaii, the most convenient mirror site is provided by the folks at RStudio (https://cloud.r-project.org/.
In brief, Windows 11 users download and install the base distribution. MacOS users must first download and install XQuartz (https://Xquartz.org), which provides the X Window System needed by R’s GUI (graphic user interface). Once XQuartz is installed, proceed to install R to your computer. MacOS users — don’t forget to drag the R.app to your Applications folder!
Start R
.
The following is a minimal look at how to use R and R Commander. Please refer to tutorials at Mike’s Workbook for Biostatistics (R work, part 1 – 10) to learn use of R and R Commander.
We’re just getting started, so the next thing to learn is how to set your working environment for your R sessions. Although we’ll discuss the R environment more as we proceed, it’s a good idea to start with a best practice action common in data science — always create and work from a working folder. This can be a local folder on your computer or pointed to a folder in your cloud storage. On my mac I usually create a folder on the desktop, example BI311, and point to that as my working folder for a project. Now, we need to point R to use the working folder — we do this at start of each R session (or modify some code R needs during start up to always point to the folder — for now, we leave that for a later exercise).
Once R is installed on your computer, start R as you would any program on your computer. For example, the icon for the R.app as it appears in the dock on a MacBook is shown in Figure 1.

Figure 1. R.app icon shown on a MacBook dock.
To point R to our working folder, there are several options. For now, we’ll go with writing and submitting a simple function in R. Recall that R has a command-line. The R prompt appears on the command line in the RGUI as the greater-than typographical symbol “>” at the beginning of a line (Fig 2). The prompt is returned by R to indicate the interpreter is ready to accept the next line of code.
First, discover where R is pointing to by submitting (type the command exactly as written then select the enter/return key) the function getwd() at the prompt. On my macbook, R returns
> getwd()
[1] "/Users/[username]/Documents"
To point to my working folder, I submit setwd("/Users/[username]/Desktop/BI311". When I run getwd() again, R returns the update
> getwd()
[1] "/Users/[username]/Desktop/BI311"
Note: Windows users will recognize that, unlike macOS and Linux, they will need to write paths with the backslash, not the forward slash. This is discussed further in Mike’s Workbook.
[username] is just a place holder here — no reason to share my actual user name!
Ready to do some work?
Where discussion requires reference to instructions on use of the R programming language, R code (instructions) the user needs to enter at the R prompt are shown in code blocks.
Courier New font within a “code block.”Until you write your own functions, the general idea is, you enter one set of commands at a time, one line at a time. For example, to create a new variable, curry.points, containing points scored by the NBA’s Steph Curry during the 2016 playoffs, type the following code at the R prompt (displayed as >, the “greater than” sign)
curry.points <- c(24,6,40,29,26,28,24,19,31,31,11,18,19).
and to obtain the mean, or arithmetic average, for curry.points at the R prompt type and enter
.
mean(curry.points).
Output from R function mean will look like the following
.
[1] 24.42857.
The R prompt appears in the RGUI as the greater-than typographical symbol “>” at the beginning of a line (Fig 2). The prompt is returned by R to indicate the interpreter is ready to accept the next line of code.
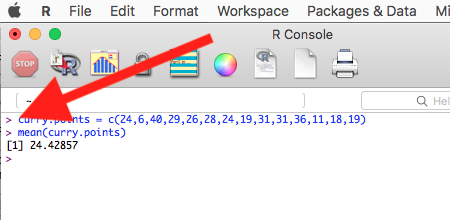
Figure 2. The R GUI on a macOS system; red arrow points to the R prompt.
Everything that exists is an object
.
A brief programmer’s note — John M. Chambers, creator of the S programming language and a member of the R-project team, once wrote that sub-header phrase about R and objects. What that means for us: programming objects can be a combination of variables, functions, and data structures. During an R session the user creates and uses objects. The ls() function is a useful R command to list objects in memory. If you have been following along with your own installed R app, then how many objects are currently available in your session of R? Answer by submitting ls(). Hint: the answer should be one object.
A routine task during analysis is to calculate an estimate then use the result in subsequent work. For example, instead of simply printing the result of mean(curry.points), we can assign the result to an object.
myResult <- mean(curry.points)
.
To confirm the new object was created, try ls() again. And, of course, there’s no particular reason to use the object name, myResult, I provided! Like any programming language, creating good object names will make your code easier to understand.
When you submit the above code, R returns the prompt, and the result of the function call is not displayed. View the result by submitting the object’s name at the R prompt, in this case, myResult. Alternatively, a simple trick is to string commands on the same line by adding ; (semicolon) at the end of the first command. For example,
myResult <- mean(curry.points); myResult
.
Write your code as script
.
While it is possible to submit code one line at a time, a much better approach is to create and manage code in a script file. A script file is just a text file with one command per line, but potentially containing many lines of code. Script files help automate R sessions. Once the code is ready, the user submits code to R from the script file.
Note: Working with scripts eliminates the R prompt, but code is still interpreted one line at a time. The user does not type the prompt in a script file.
Figure 3 shows how to create a new script file via the RGUI menu: File → New script.
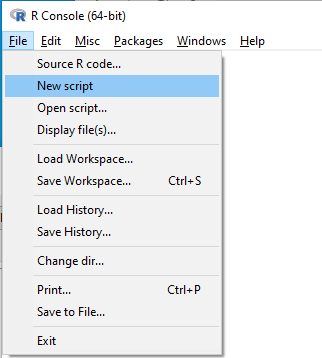
Figure 3. Screenshot of drop down menu RGUI, create new script, Windows 10.
The default text editor opens (Fig 4).
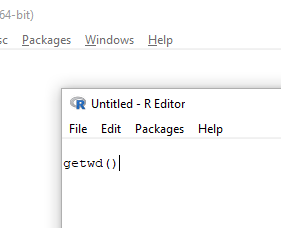
Figure 4. Screenshot of portion of R Script editor, Windows 11. A simple R command is visible.
Submit code by placing cursor at start of the code or, if code consists of multiple lines, select all of the code, then hit keyboard keys Ctrl+R (Windows 11) or for macOS, Cmd+Enter.
By default, save R script files for reuse with the file extension .R, e.g., myScript.R. Because the scripts are just text files you can use other editors that may make coding more enjoyable (see RStudio in particular, but there are many alternatives, some free to use. A good alternative is ESS).
Install R Commander package
.
By now, you have installed the base package of the R statistical programming language. The base package contains all of the components you would need to create and run data analysis and statistics on sets of data. However, you would quickly run into the need to develop functions, to write your own programs to facilitate your work. One of the great things about R is that a large community of programmers have written and contributed their own code; chances are high that someone has already written a function you would need. These functions are submitted in the form of packages. Throughout the semester we will install several R packages to extend R capabilities. R packages discussed in this book are listed at R packages of the Appendix.
Our first package to install is R Commander, Rcmdr for short. R Commander is a package that adds function to R; it provides a familiar point-and-click interface to R, which allows the user to access functions via a drop-down menu system (Fox 2017). Thus, instead of writing code to run a statistical test, Rcmdr provides a simple menu driven approach to help students select and apply the correct statistical test. R Commander also provides access to Rmarkdown and a menu approach to rendering reports.
install.packages("Rcmdr")
.
In addition, download and install the plugin
install.packages("RcmdrMisc")
See Install R Commander for detailed installation instructions.
Plugins are additional software which add function to an existing application.
Start R Commander
.
After installing Rcmdr, to start R Commander, type library(Rcmdr) at the R prompt and enter to load the library
library(Rcmdr).
On first run of R Commander you may see instructions for installing additional packages needed by R Commander. Accept the defaults and proceed to complete the installation of R Commander. Next time you start R commander the start up will be much faster since the additional packages needed by R Commander will already be present on your computer.
Note that you don’t type the R prompt and, indeed, in R Commander Script window you won’t see the prompt (Fig 4). Instead, you enter code in the R Script window, then click “Submit” button (or Win11: Ctrl+R or for macOS: Cmd+Enter), to send the command to the R interpreter. Results are sent to Output window (Fig 5).
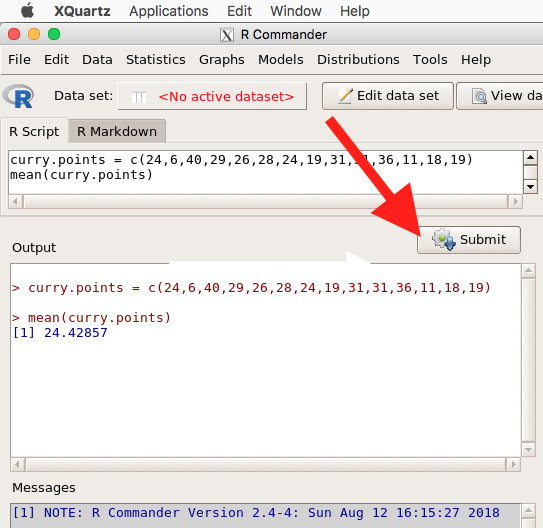
Figure 5. The windows of R Commander, macOS. From bottom to top: Messages, Output, Script (tab, Markdown) Rcmdr ver. 2.4-4.
Figure 5 shows how the R Commander GUI looks on a macOS computer. The look is similar on Microsoft Windows 11 machines (Fig 6).
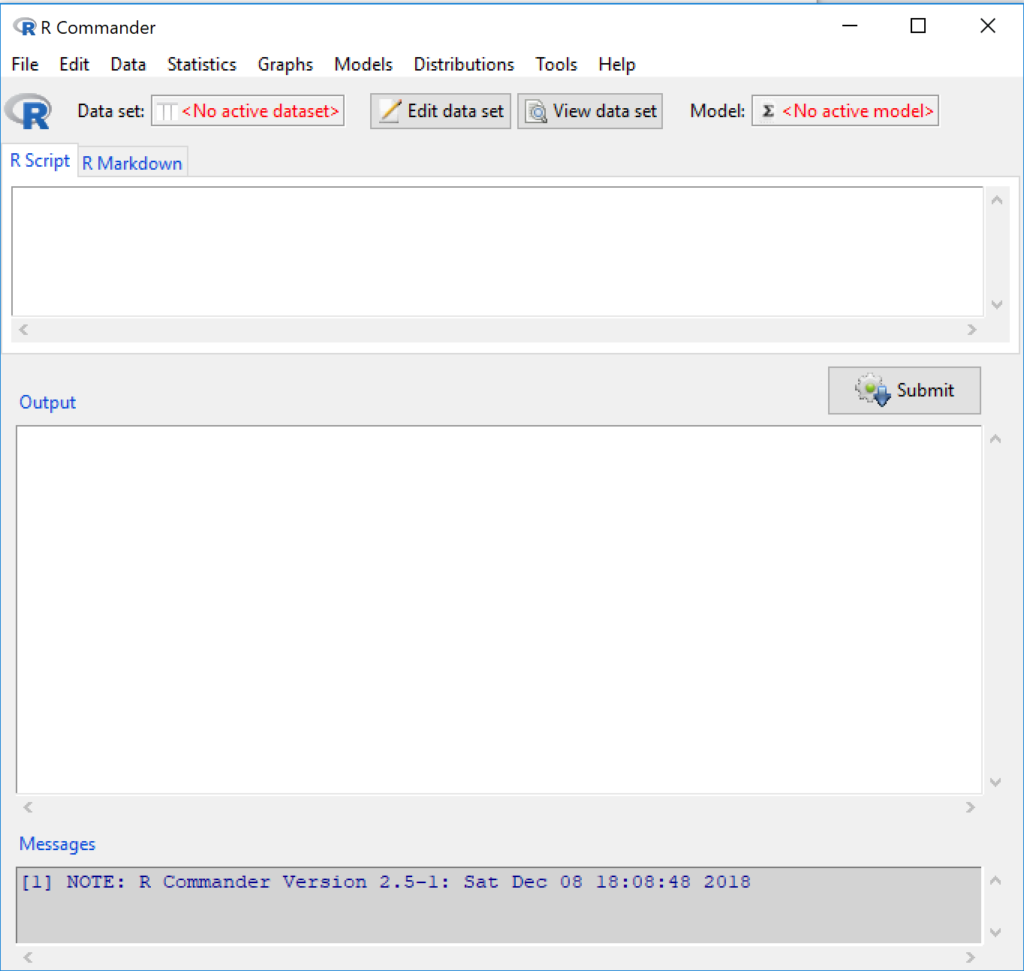
Figure 6. The windows of R Commander, Win11. From bottom to top: Messages, Output, Script (tab, R Markdown) Rcmdr ver. 2.5-1.
We use R Commander because it gives us access to code from drop-down menus, which at least initially, helps learn R (Fox 2005, Fox 2016). Later, you’ll want to write the code your self, and RStudio provides a nice environment to accomplish your data analysis.
Improve Rcmdr experience
Windows users: R Commander works best in Windows if SDI option is set. This can be accomplished during R installation (“Startup options” popup, change from default “No” to “Yes” to customize), but you can also change after installing R. Win11 users should change from MDI to SDI — from one big window to separate windows — (see Do explore settings, Figure 5). The downside of SDI is that while multiple Windows may appear during an R session, one or more windows may be hidden by an open window. For example, plots will popup in a new window and may be obscured by the Rcmdr window if full screen. A simple trick to view active windows on Win11 is to use the keyboard shortcut Alt+Tab to view and cycle between windows. What about SDI and RStudio? RStudio experience will be similar whether MDI (default) or SDI was selected.
macOS users: To improve R Commander performance, turn off Apple’s app nap (see Do explore settings, Figures 6 & 7), which should improve a Mac user’s experience with R Commander and other X Window applications.
Complete R setup by installing LaTeX and pandoc for Markdown
.
LaTeX is a system for document preparation. pandoc is a document converter system. Markdown is a language used to create formatted writing from simple text code. Once these supporting apps are installed, sophisticated reports can be generated from R sessions, by-passing copy and paste methods one might employ. See Install R Commander for instructions to add these apps.
Note: If you successfully installed R and are running R Commander, but may be having problems installing pandoc or LaTeX, then this note is for you. While there’s advantages to getting pandoc etc working, it is not essential for BI311 work.
Assuming you have Rcmdr and RcmdrMisc installed, and if you have started Rcmdr and have it up and running, then we can skip pandoc and LaTeX installation and use features of your browser to save to pdf.
R Markdown by default will print to a web page (an html document called RcmdrMarkdown.html) and display it in your default browser. To meet requirements of BI311 — you submit pdf files — we can print the html document generated from “Generate Report” in R Commander to a pdf.
- Chrome browser, right click in the web page, from the popup menu select Print, then change destination to Save as pdf.
- Safari browser, right click then select Print page (or if an option, Save page as pdf), then find at lower left find PDF and option to Save as PDF.
R Markdown
.
Markdown is a syntax for plain text formatting and is really helpful for generating clean html (web) files. R Commander also helps us with our reporting. R Markdown is provided as a tab (Fig 4, 5). Provided you have also installed pandoc on your computer, you can also convert or “render” the work into other formats including pdf and epub. Unsure if your computer has pandoc installed? If you are unsure than most likely it is not installed. 😁 Rcmdr provides a quick check — go to Tools and if you see Install auxiliary software, then click on it and a link to pandoc website to find and download installation file. You can also confirm install of pandoc by opening a terminal on your computer (e.g., search “terminal” on macOS or “cmd” on Win11), then enter pandoc –version at the shell prompt. Figure 7 shows version pandoc is installed on my Win11 HP laptop.
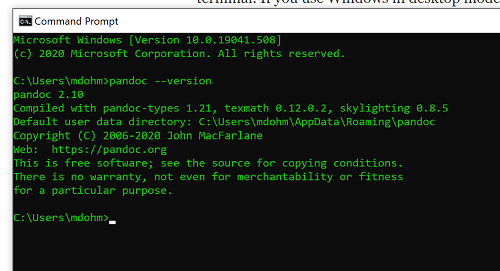
Figure 7. Screenshot of terminal window (cmd) on win11 computer, checking for installed pandoc on a win10 pc.
Enter your R code in the script window, submit your code, and your results (code, output, graphs) are neatly formatted for you by Markdown. Once the Markdown file is created in R Commander, you can then export to an html file for a a web browser, an MS Word document, or other modes.
Do explore settings!
.
After installation, R and R Commander are ready to go. However, students are advised that a few settings may need to be changed to improve performance. For example, on Win11 PCs, R Commander recommends changing from the default MDI (Multiple Document Interface) to SDI (Single Document Interface). Check the SDI button via Edit menu, select GUI preferences menu. Click save, which will make changes to .RProfile, then exit and restart R. Check to make sure the changes have been made (Fig 8).
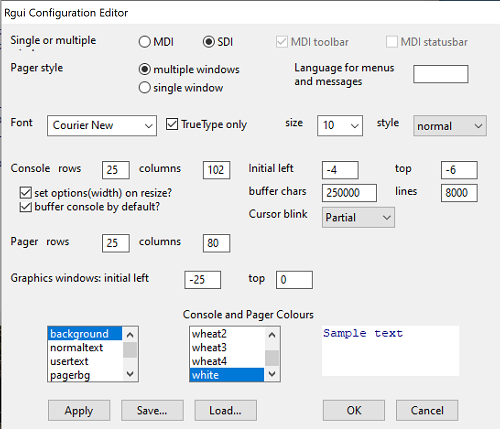
Figure 8. Screenshot of GUI preferences settings after changing from default MDI to SDI, win10
For macOS users, both R and Rcmdr will run better if you turn off Apple’s power saving feature called nap. From Rcmdr go to Tools and select Manage Mac OS X app nap for R.app… (Fig 9).
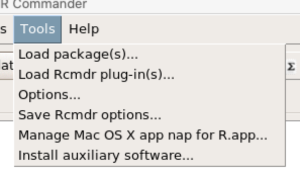
Figure 9. Screenshot Rcmdr Tools popup menu, macOS 10.15.6
A dialog box appears; select off to turn off app nap (Fig 10).
Figure 10. Screenshot Rcmdr Set app nap dialog box, macOS 10.15.6
Exit R Commander
.
Click on Rcmdr: File → Exit, then choose to exit from just R Commander, or both R Commander and R.
If you exit just R Commander or both R and R Commander, you’ll receive a pop-up request to confirm you want to quit R Commander (click yes), and a second prompt asking if you want to save your script. In general, select yes and then you’ll be able to take up where you left off. Similarly, if asked to save your workspace, choose no. If you save workspace, this creates an .RProfile text file with settings for how R and R Commander will behave the next time you start R. The file will be saved to your current working folder, which R will use the next time R starts. At least while you are getting started, you should avoid creating these .RProfile files.
As long as the current session of R is active, then the library for Rcmdr, as well as any other library loaded during the R session, is in memory. To start R Commander again while R is running, at the R prompt, type and submit
Commander()
.
Questions
.
- Biostatistics students should work through my ten R lessons, A quick look t R and R Commander, available in Mike’s Workbook for Biostatistics.
- Students should also search Internet for R tutorials and R Commander tutorials. Find recent tutorials and work through several of them. We get better when we practice.
Quiz
Chapter 1 contents
.
- Getting started
- A quick look at R and R Commander
- Chapter 1 – References
Preface
Overview
.
The following pages, loosely called Mike’s Biostatistics Book, contain the extended versions of my lectures for BI311 Biostatistics, a biology course I teach at Chaminade University.
The companion site, Mike’s Workbook for Biostatistics, provides homework, problems and projects to learn-by-doing biostatistics, and several R tutorials. (I’ve also put together a number of handouts, background, and bioinformatics support at Mike’s Genetics & Genomics Workbook, consisting of materials used to support my lab classes these past 20 years.)
The Biology department faculty require biology majors to take this course. Class standing of students range from sophomores to first year graduate students.
We use and rely heavily on R, the open source “language and environment for statistical computing” (R-project [dot] org), and the R Commander package by J Fox (Fox 2005; Fox 2016). R Commander allows students to gain confidence with R commands by use of drop down menus to access functions.
The lecture notes are written from my perspective on what matters in a semester-long, first course in Biostatistics: concepts, context and practical advice along with a generous introduction to general linear models. The focus is on applied statistics with reference to other data science skill sets as needed. Concepts are illustrated by examples and multiple choice questions to “test” reader comprehension. Examples in Mike’s Biostatistics Book are generally from real data sets in biological or biomedical research. Therefore, context and practice comes from data sets we will work on throughout the semester. The data sets are presented in the accompanying workbook, body titled Mike’s Workbook for Biostatistics.
The material presented in Mike’s Biostatistics Book provide background and the examples needed to complete the problems presented in the course workbook.
- Chapter 2: Statistical reasoning.
- Workbook: Homework 1: Assumptions.
- Chapter 3 and 4: Exploring data.
- Workbook: Homework 2A: Measurement Day results.
- Workbook: Homework 2B: Descriptive statistics.
- Chapter 5: Experimental design.
- Chapter 6: Probability.
- Workbook: Homework 3: Distributions & Probability.
- Chapter 7: Risk analysis.
- Workbook: Homework 4: Risk.
- Chapter 8: Inferential statistics.
- Workbook: Homework 5: Inference.
- Chapter 9: Qualitative (categorical) analyses.
- Workbook: Homework 6: Chi-square problems.
- Chapter 10 – 19: Quantitative (continuous) analyses.
- Chapter 10, 12, 13 – Workbook: Homework 7: t-tests and ANOVA.
- Workbook: Homework 8: Multiway ANOVA.
- Workbook: Homework 9: Correlation and simple linear regression.
- Workbook: Homework 10: Multiple linear regression.
- Chapter 11: Power analysis.
- Chapter 15: Nonparametric tests.
- Chapter 19: Distribution-free methods.
- Chapter 20: Additional topics (partial listing).
- growth curves, dose response.
- logistic regression.
- others.
- Statistical tables.
- Appendix
While the intention is to downplay lists of statistical tests in favor of developing statistical reasoning, many of the kinds of tests one comes across are introduced and discussed in this book. The intent is to introduce these tests as special cases of general (or generalized) models from a data analyst’s point of view. Think of “k-means clustering“, “independent sample t-test“, “ANOVA“, “linear regression“, and the other tests as vocabulary. We understand biology best when we can talk the talk, and the same holds for learning statistics.
The book does not include machine learning — systems that can learn and help make decisions from data — and as of September 2023, includes only a short discourse on clustering and dimensionality reduction of data sets.
About this book (and website)
.
This text book is an eBook and will undergo updates to correct errors, improve clarity, and even new content, through out the semester. Although in spirit I welcome comments, corrections, and suggestions to improve coverage and content, I have closed comments at this site to minimize spam.
Typesetting
.
The manuscript was type set from html with Markup language — See Part 09: Making a report at Mike’s Biostatistics Book — and Ghostwriter (version 2 [dot] 1 [dot] 1), and the eBook (pdf version coming soon!) was generated and edited with Calibre (version 5 [dot] 35 [dot] 0).
Hosting
Both books, Mike’s Biostatistics Book and Mike’s Workbook for Biostatistics, are hosted on my website. This site, Mike’s Biostatistics Book, is solely supported by me and I don’t monetize the site. As of Fall 2023, the site cookie only stores temporary session information during a users visit.
Equations
.
Equations in the eBook were created with LaTeX — a software system used to prepare and format documents — and saved as PNG images, or embedded in text (QuickLaTeX WordPress plug-in). Pages with many images or equations may be slow to load in your browser: in general, to improve browsing experience reduce the number of open tabs and use of additional apps.
References and citations
.
Introductory text books often lack in-line citations. The absence of in-line citations improves readability, but at the very real expense of giving credit to the original and to providing the reader the opportunity to verify facts and opinions presented. I have tried a balance: first, I included in-line citations to references. One of many remaining tasks for me to improve the book is to complete linkage of in-page citations to reference lists. I have, however, refrained from an exhaustive, dissertation-like reference listing for each point raised in the book. Second, most citations are to open access articles (or articles with pdfs available by judicious search), with the justification of the reader has access to the original material. However, this approach is a form of citation bias. Thus, I refer to reference pages as References and Suggested Readings, and don’t claim Mike’s Biostatistics Book as an authoritative voice on the subject (cf. discussion in Greenberg 2009 Bmj 339).
A note about me
.
I’m an Associate Professor of Biology at Chaminade University. My PhD was not in statistics, I trained in evolutionary physiology and quantitative genetics. Quantitative genetics is an applied field that depends heavily on use of mathematics and statistics, particularly linear models. I took courses in applied statistics while at the University of Wisconsin, but I would not call my training in statistics thorough or complete. Much of what I know comes from self-study. My strengths in biostatistics, I believe, are in translation of sometimes dense mathematics to direct use and application. Thus, I have developed a direct style to the material that I hope you will find helpful as you work on the material. It also means we won’t spend a lot of time with proofs, not because these are unimportant, but because they can side-track from developing your statistical thinking when it comes to data analysis — but primarily because this is not my strength. References are presented in the book to support the algorithms and mathematical foundations of biostatistics and to back claims I make about applied statistics.
Thus, I don’t claim that I have all of the answers, nor am I saying that the mathematical foundations are unimportant, far from it. But we have to start somewhere and I elect to spend our time on the concepts in statistics, the why do we do it this way, as opposed to the mechanics of the mathematics, the how do we do it.
What I have learned about statistics comes from publications from many real statisticians; Mike’s Biostatistics Book, such as it is, stands on on their work. I apologize in advance to any author whose work has not been given proper credit. Mistakes or mischaracterizations are, of course, mine alone.
Other sources of expertise
.
Mike’s Biostatistics Book of lecture notes is intended to provide students with a foundation in biostatistics: the concepts of assumptions, probability, sampling, description, and modeling that support a researcher’s ability to advance knowledge in biology. But you will very much benefit from other opinions, other voices. And, as much as I have adopted the online presence, it is hard to beat a book in hand as a guide. Good statistics books retain their value well passed their publication date. Some of the textbooks I have found useful over the years include the following
Introduction and general statistics
.
Chaterjee S, Price B (1977) Regression analysis by example. Wiley Interscience (5th edition now published in 2006)
Glover T, Mitchell K (2008) Introduction to biostatistics, 2nd edition. Waveland Press
Norman GR, Streiner DL (2003) PDQ Statistics, 3rd edition. BC Decker
Snedecor GW, Cochran WG (1989) Statistical methods, 8th edition. Iowa State University Press
Sokal RR, Rohlf (1981) Biometry, 2nd edition. WH Freeman (4th edition published in 2011)
Whitlock MC, Schluter D (2008) The analysis of biological data. Roberts and Company
Zar J (1999) Biostatistical analysis, 4th edition. Prentice Hall (5th edition published in 2011)
Intermediate and advanced books
Abelson RP (1995) Statistics as principled argument. Taylor & Francis (epub available)
Bulmer MG (1967) Principles of statistics. Dover Publications (epub available)
Davidson AC, Hinkley DV (1997) Bootstrap methods and their application. Cambridge University Press (epub available)
Edwards AWF (1992) Likelihood, expanded edition. Johns Hopkins University Press
Fisher RA (1934) Statistical methods for research workers, 5th edition. Oliver and Boyd (The last edition was the 14th)
Härdle W, Simar L (2003) Applied multivariate statistical analysis. Springer-Verlag
Lee PM (1989) Bayesian statistics: An introduction. Oxford University Press – McCullagh P, Nelder JA (1989) Generalized linear models, 2nd edition. Chapman and Hall
Montgomery DC, Peck EA (1992) Introduction to linear regression analysis, 2nd edition. John Wiley & Sons (5th edition published in 2013)
Neter J, Wasserman W, Kutner MH (1989) Applied linear regression models, 2nd edition. Robert D Irwin (4th edition published in 2003)
Quinn GP, Keough MJ (2002) Experimental design and data analysis for biologists. Cambridge University Press.
Shao J (2003) Mathematical statistics, 2nd ed. Springer Science
Wei WWS (1990) Time series analysis. Addison-Wesley (2nd edition published in 2005)
Some of these titles are old!
.
One of the good things about statistics is that many of the standard statistical applications were developed a long time ago, so “old textbooks” in statistics retain their value. A quick search online will result in many options to purchase one or more of these books for under $10. In addition, most of the books listed above have new editions; where appropriate I have listed the most recent available edition.
What about books on R?
None of the listed books teach R. Between Mike’s Biostatistics Book and the companion Mike’s Workbook for Biostatistics, several tutorial and lots of worked examples are provided to help you learn how to use R to help statistical work. A quick Google search, e.g., “free online books learn R,” returns thousands of suggested titles. Search “R tutorials,” for millions more. Chances are, if you have a question about how to do some task in R, someone has already solved the task and published code examples for you to borrow (always cite your sources!).
Concluding remarks about these lecture notes
.
These collected lecture notes will serve as your official textbook – I have tried to make them accurate, informative, and yet balanced between providing too much detail while still providing depth to the presentation. In class, lecture slides will be provided as outline to these more extensive notes. Homework and quizzes support the progress through the notes.
The lecture notes contained in Mike’s Biostatistics Book are very much a work in progress, with some areas more developed than others. If you find areas that make no sense, seem abrupt, or you would like more examples, please do let me know. Your input is important to improve this textbook; the Discussion Forum on the course website is a good place to do lend your critiques and suggestions.
Like most subjects one voice is not enough; you will benefit from acquiring a second opinion, either from one or more of the books listed above or from the many online sites on statistics you will find. The good news is that you will find substantial overlap between what I write and other sources you may acquire because the topics we will cover are foundational and my take is mainstream.
However, you will also find some differences in detail. For one example, I have included much more on risk analysis and an epidemiology tilt as compared to many of the the titles listed under the Introduction and General statistics category. For a second example, I give a different perspective on how to work with probability calculations, emphasizing use of natural numbers over frequency calculations. Many examples provided in the book are drawn from data sets created in lab classes you are or will take while you are at Chaminade University: growth curves, dose-response, working with RT-PCR traces, multi-well plate assays and more.
Preface contents
.
Preface