1.1 – A quick look at R and R Commander
A first look at R.
R is a programming language for statistical computing (Venables et al 2009). R is an interpreted language — when you run (execute) an R program — the R interpreter intercepts and runs the code immediately through its command-line interface, one line at time. Python is another popular interpreted language common in data science. Interpreted languages are in contrast to compiled languages, like C++ and Rust, where program code is sent to a compiler to a machine language application. R is free to use and available via many platforms, from local installations on a person’s laptop to running on cloud servers (Fig 1). 
Figure 1. Choose how you will interact with R: on your computer (Blue box) or in the cloud (Red box). Users of Linux, macOS, and WinPC can either install R to their computer or choose to access R in the cloud (and/or, black arrow).
Before you begin, you have a choice: how do you want to work with R (Fig 1)? In part, the choice depends on your computing device. If you have a desktop or laptop computer, running Linux, Apple macOS, or Microsoft Windows, then R can be installed onto your computer (blue box, Fig 1). If, however, you have a ChromeBook, Apple iPad or iPhone, or any Android Tablet or Smartphone, you would go with the second option (red box, Fig 1).
For those going with installing R to their computer, the second choice is to pick the environment for creating and running scripts and managing file input and output during a session. Working environments for R in the cloud include RStudio and Jupyter Notebooks (eg, Google CoLab) (red box, Fig 1 — see Note 2 below). For installed R, two suggested environments are R Commander and RStudio, an Integrated Development Environment or IDE, RStudio (Fig 1). The choice depends on your needs — an IDE is helpful when you plan to write a lot of code, it’s a comprehensive programming environment, which includes many tools that support writing code from scratch.
For our course at Chaminade University, I recommend R Commander because of the drop-down menu approach to accessing statistical functions in R; R Commander is not a comprehensive environment, but it provides the necessary scripting window along with access to markdown editing. R Commander, Rcmdr, is an R package that provide a graphical user interface or GUI for R. It is based on the tcltk package. R must be installed first before installing the R Commander package. R Commander provides familiar menu functions, like many commercial statistical software. R Commander, like the IDE RStudio, is not necessary for R programming or conducting statistical analysis or even generating reports via markdown. R Commander, however, shrinks the learning curve with R programming basics so that students can focus on learning statistics, which is the chief objective of the Biostatistics course at Chaminade.
The following steps user through use of R and R Commander, from installation to writing and running commands. Mike’s Workbook for Biostatistics has a ten-part tutorial, A quick look at R and R Commander, which I recommend.
Note 1: Getting started? By all means rely on Mike’s Biostatistics Book (Appendix: Install R, Install R Commander, Use R in the cloud) and blogs or other online tutorials to point you in the right direction. You’ll also find many free and online books that may provide the right voice to get you working with R. However, the best way to learn is to go to the source. The R team has provided extensive documentation, all included as part of your installation of R. In R, run the command RShowDoc("doc name"). replace doc name with the name of the R manual or R user guide. For example, the Venables publication is accessed as RShowDoc("R-intro"). Similarly, the manual for installation is RShowDoc("R-admin") and the manual for R data import/export is RShowDoc("R-data").
Install R.
.
Full installation instructions are available at Install R, and for the R Commander package, at Install R Commander. Here, we provide a brief overview of the installation process.
Note 2: The instructions at Mike’s Biostatistics Book assume use of R on a personal computer running updated Microsoft Windows or Apple macOS operating systems. For Linux instructions, eg, Ubuntu distro, see How to install R on Ubuntu 22.04. For Chromebook users, if you can install a Linux subsystem, then you can also install and run R although it’s not a trivial installation. For instructions to install R see Levi’s excellent write up at levente.littvay.hu/chromebook/. (This works best with Intel-based CPUs — see my initial attempts with an inexpensive Chromebook at Install R.)
Another option is to run R in the cloud via service like Google’s Colab or CoCalc hosted by SageMath. Both support Jupyter Notebooks, a “web-based interactive computational environment.” Neither cloud-based service supports use of R Commander (because R Commander interacts with your local hardware). Colab is the route I’d choose if I don’t have access to a local installation of R. See Use R in the cloud for more details.
Download a copy of the R installation file appropriate for your computer from one of the Comprehensive R Archive Network (CRAN) mirror site of the r-project.org. For Hawaii, the most convenient mirror site is provided by the folks at RStudio (https://cloud.r-project.org/. In brief, Windows 11 users download and install the base distribution. MacOS users must first download and install XQuartz (https://Xquartz.org), which provides the X Window System needed by R’s GUI (graphic user interface). Once XQuartz is installed, proceed to install R to your computer. MacOS users — don’t forget to drag the R.app to your Applications folder!
Start R.
.
The following is a minimal look at how to use R and R Commander. Please refer to tutorials at Mike’s Workbook for Biostatistics (R work, part 1 – 10) to learn use of R and R Commander.
We’re just getting started, so the next thing to learn is how to set your working environment for your R sessions. Although we’ll discuss the R environment more as we proceed, it’s a good idea to start with a best practice action common in data science — always create and work from a working folder. This can be a local folder on your computer or pointed to a folder in your cloud storage. On my mac I usually create a folder on the desktop, example BI-311, and point to that as my working folder for a project. Now, we need to point R to use the working folder — we do this at start of each R session (or modify some code R needs during start up to always point to the folder — for now, we leave that for a later exercise).
Once R is installed on your computer, start R as you would any program on your computer. For example, the icon for the R.app as it appears in the dock on a MacBook is shown in Figure 2.

Figure 2. R.app icon shown on a MacBook dock.
To point R to our working folder, there are several options. For now, we’ll go with writing and submitting a simple function in R. Recall that R has a command-line. The R prompt appears on the command line in the RGUI as the greater-than typographical symbol “>” at the beginning of a line (Fig 2). The prompt is returned by R to indicate the interpreter is ready to accept the next line of code.
First, discover where R is pointing to by submitting (type the command exactly as written then select the enter/return key) the function getwd() at the prompt. On my MacBook, R returns
> getwd()
[1] "/Users/[username]/Documents"
To point to my working folder, I submit setwd("/Users/[username]/Desktop/BI311". When I run getwd() again, R returns the update
> getwd()
[1] "/Users/[username]/Desktop/BI311"
Note 3: Windows users will recognize that, unlike macOS and Linux, they will need to write paths with the backslash, not the forward slash. This is discussed further in Mike’s Workbook.
[username] is just a placeholder here — no reason to share my actual user name!
Ready to do some work?
R coding practices in this book and in the companion workbook roughly follow guidance outlined in a September 2018 R-Bloggers post, “R Code — Best practices,” by The R Trader. Of note, naming conventions: variables are nouns, variable names are written in lowerCamelCase; functions are verbs, function names are period separated, eg, my.function; script file names all end with .R and are written snake_case.R. Comments, preceded by # and a single space, eg, # A brief comment, are sparingly included in code, but reserved for explanatory sentences in the text. We use <- and not = for assignment statements.
Where discussion requires reference to instructions on use of the R programming language, R code (instructions) the user needs to enter at the R prompt are shown in code blocks.
Courier New font within a “code block.”Until you write your own functions, the general idea is, you enter one set of commands at a time, one line at a time. For example, to create a new variable, curryPoints, containing points scored by the NBA’s Steph Curry during the 2016 playoffs, type the following code at the R prompt (displayed as >, the “greater than” sign)
curryPoints <- c(24,6,40,29,26,28,24,19,31,31,11,18,19).
and to obtain the mean, or arithmetic average, for curryPoints at the R prompt type and enter
.
mean(curryPoints).
Output from R function mean will look like the following
.
[1] 24.42857.
Functions in R, or any other data science programming language, are intended to automate a task.
Note 4: A good data management habit — store the variable as an R object called a data.frame.
df.curryPoints <- data.frame(curryPoints)
The R prompt appears in the RGUI as the greater-than typographical symbol “>” at the beginning of a line (Fig 3). The prompt is returned by R to indicate the interpreter is ready to accept the next line of code.
![Figure 1. The R GUI on a macOS system; red arrow points to the R prompt. Screenshot of the R GUI on a macOS system; red arrow points to the R prompt. The visible code reads curry.points and some numbers between parentheses, next line calls for the mean of the object, and the last line begins with [1] and the result.](https://biostatistics.letgen.org/wp-content/uploads/2020/09/RGUI_prompt.png)
Figure 3. The R GUI on a macOS system; red arrow points to the R prompt.
Note 5: We just demonstrated one of several ways data can be brought into an R session, by creating a data.frame directly from a vector. Other methods include
- Reading a text file from your computer, eg, CSV, by
read.csv() - Importing a spreadsheet (Excel file) from your computer
using readxl::read_excel() - Entering small datasets directly using
read.table()with thetext= argument
Everything that exists is an object.
.
A brief programmer’s note — John M. Chambers, creator of the S programming language and a member of the R-project team, once wrote that sub-header phrase about R and objects. What that means for us: programming objects can be a combination of variables, functions, and data structures. During an R session the user creates and uses objects. The ls() function is a useful R command to list objects in memory. If you have been following along with your own installed R app, then how many objects are currently available in your session of R? Answer by submitting ls(). Hint: the answer should be one object.
A routine task during analysis is to calculate an estimate then use the result in subsequent work. For example, instead of simply printing the result of mean(curryPoints), we can assign the result to an object.
myResult <- mean(curryPoints)
.
To confirm the new object was created, try ls() again. And, of course, there’s no particular reason to use the object name, myResult, I provided! Like any programming language, creating good object names will make your code easier to understand.
When you submit the above code, R returns the prompt, and the result of the function call is not displayed. View the result by submitting the object’s name at the R prompt, in this case, myResult. Alternatively, a simple trick is to string commands on the same line by adding ; (semicolon) at the end of the first command. For example,
myResult <- mean(curryRoints); myResult
.
Objects created during an R session, like curryPoints, are stored in memory within the Global Environment. To remove an object, use the command rm(myVariable). Why remove objects during an R session? If you are attempting multiple tasks in a homework assignment, removing unneeded objects helps maintain a clean and organized workspace, improving code readability and reducing the chance of accidental errors.
Write your code as script.
.
While it is possible to submit code one line at a time, a much better approach is to create and manage code in a script file. A script file is just a text file with one command per line, but potentially containing many lines of code. Script files help automate R sessions. Once the code is ready, the user submits code to R from the script file.
Note 6: Working with scripts eliminates the R prompt, but code is still interpreted one line at a time. The user does not type the prompt in a script file.
Figure 4 shows how to create a new script file via the RGUI menu: File → New script.

Figure 4. Screenshot of drop down menu RGUI, create new script, Windows 11.
The default text editor opens (Fig 5).
Figure 5. Screenshot of portion of R Script editor, Windows 11. A simple R command is visible.
Submit code by placing cursor at start of the code or, if code consists of multiple lines, select all of the code, then hit keyboard keys Ctrl+R (Windows 11) or for macOS, Cmd+Enter.
I recommend starting with a clean environment, by using rm(list = ls()) at the beginning of a script.
By default, save R script files for reuse with the file extension .R, eg, my_script.R. Because the scripts are just text files you can use other editors that may make coding more enjoyable (see RStudio in particular, but there are many alternatives, some free to use. A good alternative is ESS).
Install R Commander package.
.
By now, you have installed the base package of the R statistical programming language (see for detailed instructions). The base package contains all of the components you would need to create and run data analysis and statistics on sets of data. However, you would quickly run into the need to develop functions, to write your own programs to facilitate your work. One of the great things about R is that a large community of programmers have written and contributed their own code; chances are high that someone has already written a function you would need. These functions are submitted in the form of packages. Throughout the semester we will install several R packages to extend R capabilities. R packages discussed in this book are listed at R packages of the Appendix.
Our first package to install is R Commander, Rcmdr for short. R Commander is a package that adds function to R; it provides a familiar point-and-click interface to R, which allows the user to access functions via a drop-down menu system (Fox 2017). Thus, instead of writing code to run a statistical test, Rcmdr provides a simple menu driven approach to help students select and apply the correct statistical test. R Commander also provides access to Rmarkdown and a menu approach to rendering reports.
install.packages("Rcmdr")
.
In addition, download and install the plugin
install.packages("RcmdrMisc")
See Install R Commander for detailed installation instructions.
Note 7: Plugins are additional software which add function to an existing application.
Start R Commander.
.
After installing Rcmdr, to start R Commander, type library(Rcmdr) at the R prompt and enter to load the library
library(Rcmdr).
On first run of R Commander you may see instructions for installing additional packages needed by R Commander. Accept the defaults and proceed to complete the installation of R Commander. Next time you start R commander the start up will be much faster since the additional packages needed by R Commander will already be present on your computer.
Note that you don’t type the R prompt and, indeed, in R Commander Script window you won’t see the prompt (Fig 6). Instead, you enter code in the R Script window, then click “Submit” button (or Win11: Ctrl+R or for macOS: Cmd+Enter), to send the command to the R interpreter. Results are sent to Output window (Fig 6).
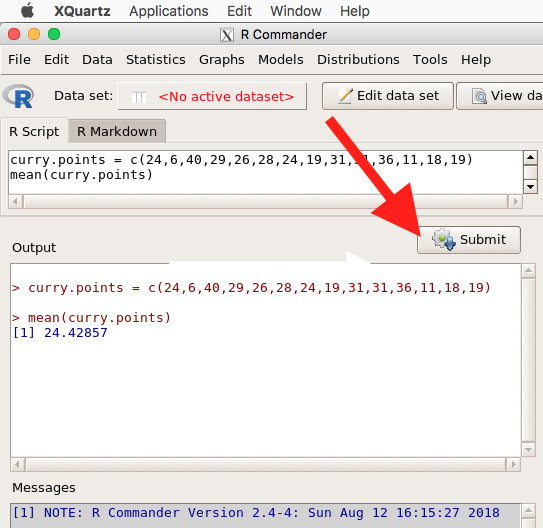
Figure 6. The windows of R Commander, macOS. From bottom to top: Messages, Output, Script (tab, Markdown) Rcmdr ver. 2.4-4.
Figure 6 shows how the R Commander GUI looks on a macOS computer. The look is similar on Microsoft Windows 11 machines (Fig 7).
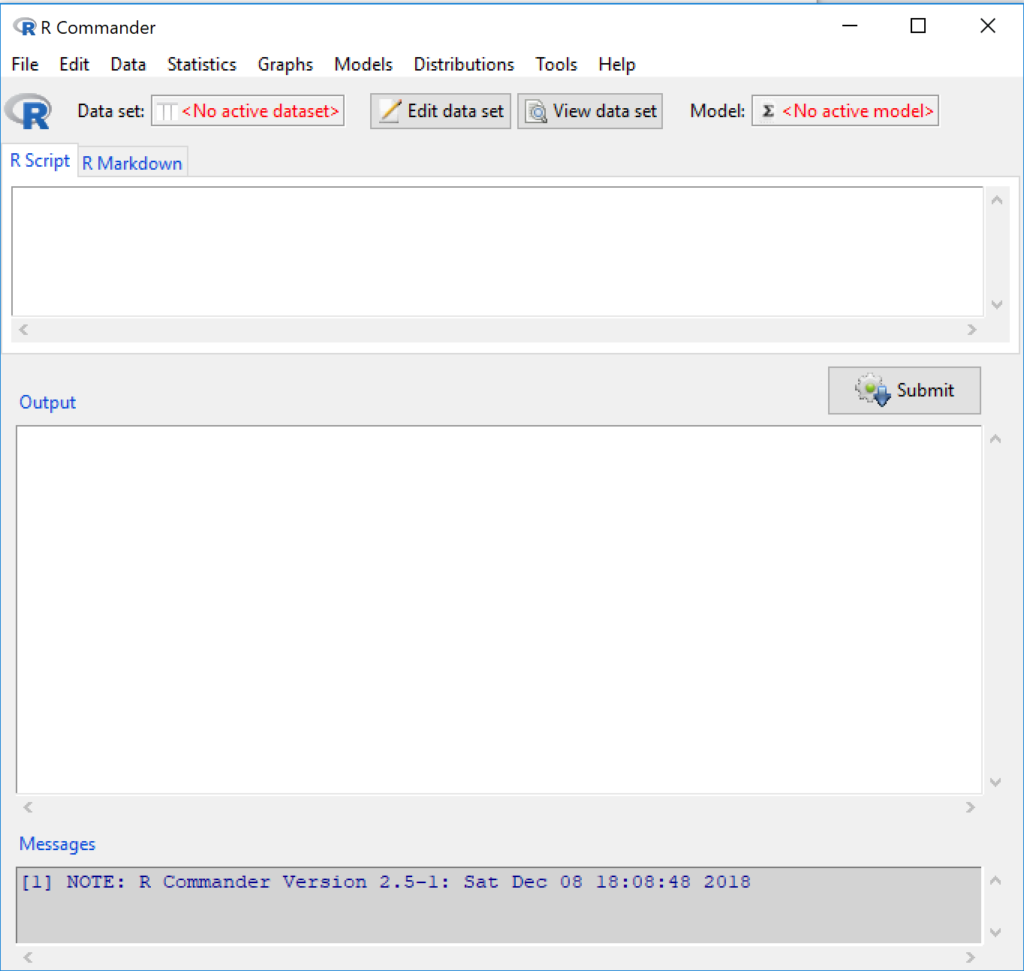
Figure 7. The windows of R Commander, Win11. From bottom to top: Messages, Output, Script (tab, R Markdown) Rcmdr ver. 2.5-1.
We use R Commander because it gives us access to code from drop-down menus, which at least initially, helps learn R (Fox 2005, Fox 2016). Later, you’ll want to write the code yourself, and RStudio provides a nice environment to accomplish your data analysis.
Improve Rcmdr experience.
Windows users: R Commander works best in Windows if SDI option is set. This can be accomplished during R installation (“Startup options” popup, change from default “No” to “Yes” to customize), but you can also change after installing R. Win11 users should change from MDI to SDI — from one big window to separate windows — (see Do explore settings, WinPC see Fig 9; macOS see Fig 10). The downside of SDI is that while multiple Windows may appear during an R session, one or more windows may be hidden by an open window. For example, plots will popup in a new window and may be obscured by the Rcmdr window if full screen. A simple trick to view active windows on Win11 is to use the keyboard shortcut Alt+Tab to view and cycle between windows. What about SDI and RStudio? RStudio experience will be similar whether MDI (default) or SDI was selected.
macOS users: To improve R Commander performance, turn off Apple’s app nap (see Do explore settings, Figures 6), which should improve a Mac user’s experience with R Commander and other X Window applications.
Complete R setup by installing LaTeX and pandoc for Markdown.
.
LaTeX is a system for document preparation. pandoc is a document converter system. Markdown is a language used to create formatted writing from simple text code. Once these supporting apps are installed, sophisticated reports can be generated from R sessions, by-passing copy and paste methods one might employ. See Install R Commander for instructions to add these apps.
Note 8: If you successfully installed R and are running R Commander, but may be having problems installing pandoc or LaTeX, then this note is for you. While there’s advantages to getting pandoc etc working, it is not essential for BI311 work.
Assuming you have Rcmdr and RcmdrMisc installed, and if you have started Rcmdr and have it up and running, then we can skip pandoc and LaTeX installation and use features of your browser to save to pdf.
R Markdown by default will print to a web page (an html document called RcmdrMarkdown.html) and display it in your default browser. To meet requirements of BI311 — you submit pdf files — we can print the html document generated from “Generate Report” in R Commander to a pdf.
- Chrome browser, right click in the web page, from the popup menu select Print, then change destination to Save as pdf.
- Safari browser, right click then select Print page (or if an option, Save page as pdf), then find at lower left find PDF and option to Save as PDF.
R Markdown.
.
Markdown is a syntax for plain text formatting and is really helpful for generating clean html (web) files. R Commander also helps us with our reporting. R Markdown is provided as a tab (Fig 6, Fig 7). Provided you have also installed pandoc on your computer, you can also convert or “render” the work into other formats including pdf and epub. Unsure if your computer has pandoc installed? If you are unsure than most likely it is not installed. 😁 Rcmdr provides a quick check — go to Tools and if you see Install auxiliary software, then click on it and a link to pandoc website to find and download installation file. You can also confirm install of pandoc by opening a terminal on your computer (eg, search “terminal” on macOS or “cmd” on Win11), then enter pandoc –version at the shell prompt. Figure 8 shows version pandoc is installed on my Win11 HP laptop.
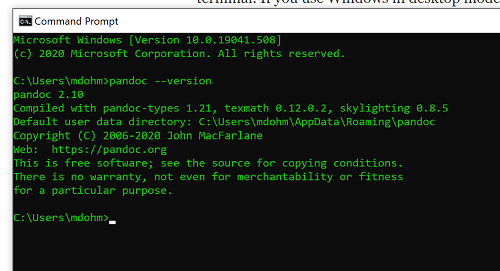
Figure 8. Screenshot of terminal window (cmd) on win11 computer, checking for installed pandoc on a win10 pc.
Enter your R code in the script window, submit your code, and your results (code, output, graphs) are neatly formatted for you by Markdown. Once the Markdown file is created in R Commander, you can then export to an html file for a a web browser, an MS Word document, or other modes.
Do explore settings!
.
After installation, R and R Commander are ready to go. However, students are advised that a few settings may need to be changed to improve performance. For example, on Win11 PCs, R Commander recommends changing from the default MDI (Multiple Document Interface) to SDI (Single Document Interface). Check the SDI button via Edit menu, select GUI preferences menu. Click save, which will make changes to .RProfile, then exit and restart R. Check to make sure the changes have been made (Fig 9).
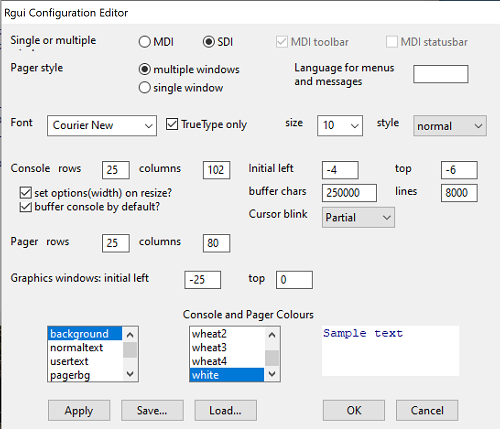
Figure 9. Screenshot of GUI preferences settings after changing from default MDI to SDI, win10
For macOS users, both R and Rcmdr will run better if you turn off Apple’s power saving feature called nap. From Rcmdr go to Tools and select Manage Mac OS X app nap for R.app… (Fig 10).
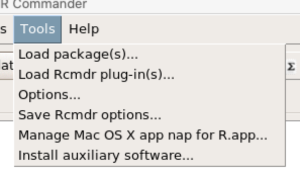
Figure 10. Screenshot Rcmdr Tools popup menu, macOS 10.15.6
A dialog box appears; select off to turn off app nap (Fig 11).
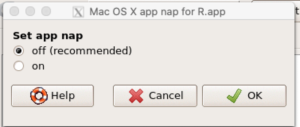
Figure 11. Screenshot Rcmdr Set app nap dialog box, macOS 10.15.6
Exit R Commander.
.
Click on Rcmdr: File → Exit, then choose to exit from just R Commander, or both R Commander and R.
If you exit just R Commander or both R and R Commander, you’ll receive a pop-up request to confirm you want to quit R Commander (click yes), and a second prompt asking if you want to save your script. In general, select yes and then you’ll be able to take up where you left off. Similarly, if asked to save your workspace, choose no. If you save workspace, this creates an .RProfile text file with settings for how R and R Commander will behave the next time you start R. The file will be saved to your current working folder, which R will use the next time R starts. At least while you are getting started, you should avoid creating these .RProfile files.
As long as the current session of R is active, then the library for Rcmdr, as well as any other library loaded during the R session, is in memory. To start R Commander again while R is running, at the R prompt, type and submit
Commander()
Questions.
- Biostatistics students should work through my ten R lessons, A quick look t R and R Commander, available in Mike’s Workbook for Biostatistics.
- Students should also search Internet for R tutorials and R Commander tutorials. Find recent tutorials and work through several of them. We get better when we practice.
Quiz Chapter 1.1
A quick look at R and R Commander
Chapter 1 contents
.
- Getting started
- A quick look at R and R Commander
- Chapter 1 – References